Stature estimation a la Bayes

We will use OpenBUGS to estimate the stature, but OpenBUGS is "hungry." So first we will have to "feed" OpenBUGS some numbers from R.
# from: https://github.com/cran/corpcor/blob/master/R/smtools.R
vec2sm = function(vec, diag = FALSE, order = NULL)
{
# dimension of matrix
n = (sqrt(1+8*length(vec))+1)/2
if (diag == TRUE) n = n-1
if ( ceiling(n) != floor(n) )
stop("Length of vector incompatible with symmetric matrix")
# fill lower triangle of matrix
m = matrix(NA, nrow=n, ncol=n)
lo = lower.tri(m, diag)
if (is.null(order))
{
m[lo] = vec
}
else
{
# sort vector according to order
vec.in.order = rep(NA, length(order))
vec.in.order[order] = vec
m[lo] = vec.in.order
}
# symmetrize
for (i in 1:(n-1))
for (j in (i+1):n)
m[i, j] = m[j, i]
return( m )
}
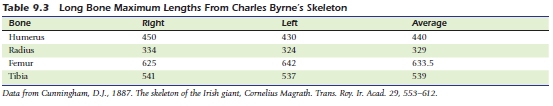
giant=c(440,329,633.5,539)
# means
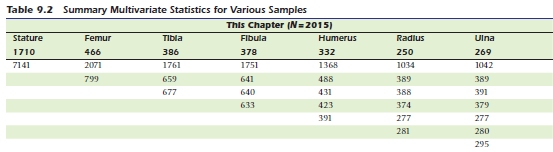
mu = c(1710,332,250,466,386)
mux = mu[1]
muy = mu[-1]
# Upper triangular part of variance-covariance matrix packed as a vector
v=c(7141,2071,1761,1368,1034,799,659,488,389,677,431,388,391,277,281)
# Unpack to symmetric matrix
V=vec2sm(v,diag=T)
# Being indecisive, I changed the order of variables
V = V[c(1,4,5,2,3),c(1,4,5,2,3)]
# Partition matrix
vx = V[1,1]
vxy = V[1,-1]
Vyy = V[-1,-1]
cat('\n\n')
cat(paste('Informative prior with mean = ',round(mux,0)))
cat('\n')
cat(paste(' and precision = ',round(1/vx,5)))
cat('\n\n')
# Get regression coefficients for long bones predicted by stature
b = round(vxy/vx,7)
a = round(muy - b*mux,4)
cat('Multivariate regression coefficients: \n\n')
dput(t(rbind(a,b)))
# Inverse of conditional var/covar matrix
cat('\n\n')
cat('Inverse of residual var/covar matrix: \n\n')
dput(round(solve(Vyy-vxy%*%solve(vx)%*%t(vxy)),6))
cat('\n\n')
b=vxy%*%solve(Vyy)
a=mux-b%*%muy
stat=as.numeric(a+b%*%giant)
cat(paste('Stature estimated by multiple regression = ',round(stat,0)))
cat('\n\n')
}
get.nums()
| model { # Uniform (uninformative) prior stat[1] ~ dunif(0,10000) # N(1710, 84.5) prior from reference sample, multiple regression estimate = 2159 stat[2] ~ dnorm(1710, 0.00014) for(j in 1:4){ for(i in 1:2) {Y.pred[i,j]<-theta[j] + theta[4+j] * stat[i]}} for(i in 1:2) {Y[i,1:4] ~ dmnorm(Y.pred[i,], tau[,])} } #data, note "stuttering" of long bone data list(Y=structure(.Data=c(440,329,633.5,539,440,329,633.5,539), .Dim=c(2,4)),theta=c(4.4156, 2.396, -29.9263, -35.693, 0.1915698, 0.1447976, 0.2900154, 0.2466041), tau = structure(.Data= c(0.01405, -0.006103, -0.003993, 0.000363, -0.006103, 0.019939, 0.000473, -0.00886, -0.003993, 0.000473, 0.010499, -0.005133, 0.000363, -0.00886, -0.005133, 0.011968),.Dim=c(4,4))) |
Stature as an evidentiary problem
N = 2015
# means
mu = c(1710,332,250,466,386)
mux = mu[1]
muy = mu[-1]
# Upper triangular part of variance-covariance matrix packed as a vector
v=c(7141,2071,1761,1368,1034,799,659,488,389,677,431,388,391,277,281)
# Unpack to symmetric matrix
V=vec2sm(v,diag=T)
# Being indecisive, I changed the order of variables
V = V[c(1,4,5,2,3),c(1,4,5,2,3)]
# Partition matrix
vx = V[1,1]
vxy = V[1,-1]
Vyy = V[-1,-1]
# Get regression coefficients for long bones predicted by stature
b = vxy/vx
a = muy - b*mux
stat=1710
v.stat=84.5^2
bones.pred = b*stat+a
cat(paste('\n\nPredicted bone lengths at stature = ',stat,'\n'))
dput(round(bones.pred,4))
cat('\n\n')
C = Vyy-vxy%*%solve(vx)%*%t(vxy)
tau = solve(b%o%b*v.stat + C)
cat('Inverse var-covar matrix of predicted bones:\n')
dput(round(tau,10))
stat=2310
v.stat=25^2
bones.pred = b*stat+a
cat(paste('\n\nPredicted bone lengths at stature = ',stat,'\n'))
dput(round(bones.pred,4))
cat('\n\n')
tau = solve(b%o%b*v.stat + C)
cat('Inverse var-covar matrix of predicted bones:\n')
dput(round(tau,10))
cat('\n\n')
}
get.nums2()
| model { bones[1:4] ~ dmnorm(bones.pred[], tau[,]) Log.like<- -deviance(bones[1:4],bones[1:4])/2 } #data - for N(1710, 84.5) list(bones=c(440,329,633.5,539),bones.pred=c(332, 250, 466, 386),tau = structure(.Data= c(0.0131532986, -0.00570302, -0.0053065435, 6.0402E-05, -0.00570302, 0.0197598473, 0.0010589323, -0.0087248683, -0.0053065435, 0.0010589323, 0.0085757009, -0.0055758772, 6.0402E-05, -0.0087248683, -0.0055758772, 0.0118660013),.Dim=c(4,4))) #data - for N(2310, 25) list(bones=c(440,329,633.5,539),bones.pred=c(446.9419, 336.8786, 640.0092, 533.9625),tau = structure(.Data= c(0.0137861427, -0.0059854557, -0.0043798194, 0.0002739931, -0.0059854557, 0.0198858971, 0.0006453392, -0.0088201931, -0.0043798194, 0.0006453392, 0.0099327771, -0.0052630986, 0.0002739931, -0.0088201931, -0.0052630986, 0.0119380905),.Dim=c(4,4))) |
formatC(exp(-14.38--33.22),format='d',big.mark=',')